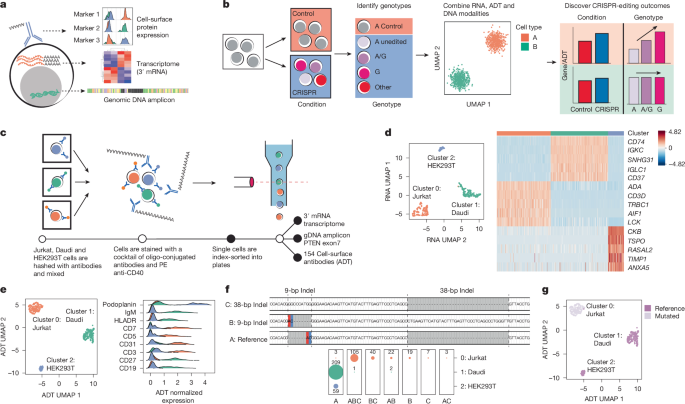
"Large-scale genome-wide association studies have identified many complex trait loci related to autoimmune, metabolic, and infectious diseases, yet assigning function to non-coding variants remains challenging."
"CRISPR-Cas technologies, particularly with base-pair resolution, offer powerful tools for genetic modifications and identifying regulatory elements, but low editing efficiency is a significant limitation."
"Current methods often assume editing efficiency is high and rely on sequencing single guide RNA as a proxy, which can lead to substantial inaccuracies in detecting effective edits."
"Single-cell data could enhance the efficiency of CRISPR screens by distinguishing between edited and non-edited cells, overcoming some limitations in current experimental setups."
Genome-wide association studies have identified numerous complex trait loci related to autoimmune, metabolic, and infectious diseases. Despite the effectiveness of computational methods in mapping signals to variants, accurately assigning function to non-coding variants poses significant challenges. Precise genomic editing using base-pair-resolution CRISPR-Cas technologies shows promise in addressing this issue. Although large-scale single-cell CRISPR screenings have identified key regulatory elements, limitations like low editing efficiency and nonspecific effects complicate functional analysis. Current methodologies rely on single guide RNA sequencing to infer editing success, which can lead to inaccuracies in distinguishing effective edits.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]